Our list of miscellaneous genetic stocks (download in *.xlsx format), now containing 2,201 accessions, was last updated in 2020 and is a revision of the previous list issued in TGC Report 62 (2012). Extinct, obsolete, or faulty accessions have been dropped. Most stocks are maintained as homozygotes and samples of 25 seed are provided upon request. However, some stocks, such as multiple marker combinations, aneuploids, and some prebreds, may be weak and relatively infertile, consequently, seed supplies may be limited. Other accessions may be temporarily unavailable during seed regeneration. Other types of stocks are described in our Monogenic Stocks list or in our Wild Species Accessions list. Additional information on these stocks is available under Database Queries/Accession Search.
The main categories of genetic stocks are described below:
Modern and Vintage Cultivars (222). We maintain a set of cultivars, inbreds, and breeding lines for various purposes, mainly as nonmutant control genotypes for specific mutants, standards for genetic comparisons, sources of disease resistances, or for other purposes. Cultivar Marglobe is considered the standard for tomato gene (mutant) nomenclature. Most lines have been maintained by selfing for many generations.
Latin American Cultivars (349). This collection of Latin-American cultivars has been assembled from various sources but principally from our collecting trips, often at local markets. With a few exceptions they are indigenous in the sense that they are not recently introduced lines. Many of them are extinct in the source region, having been replaced by modern cultivars.

S. pennellii Introgression Lines (83). This collection of ILs was developed by Eshed and Zamir (1994 Euphytica 79:175; 1999 TGC Report 49:26). Each IL is homozygous for a single introgression from S. pennellii (LA0716) in the background of S. lycopersicum cv. M-82 (LA3475). (IL 8-1 is heterozygous for a short and a long introgression.) The entire pennellii genome is thereby represented by a minimum 50 lines with overlapping introgressions. Recombinant sublines provide increased mapping resolution in some regions. (The IL 5-4 sublines are described in Jones et al. 2007 Amer. J. Bot. 94: 935 and Xu et al. 2008 Theor. Appl. Genet. 117: 221. Click here for more information.
S. habrochaites ILs (93). These ILs represent the genome of S. habrochaites LA1777 in the background of S. lycopersicum cv. E-6203 (LA4024) via homozygous chromosome segments (Monforte and Tanksley 2000 Genome 43:803). The first 57 lines (LA3913 - LA3969) represent approximately 85% of the donor genome, while the remaining lines (LA3970 - LA4010) contain different introgressions, mostly derivatives of the first group, for added mapping precision. Unlike the pennellii ILs above, each habrochaites IL may contain more than one introgression, representing one to several chromosomes, similar to the S. lycopersicoides ILs. Click here for more information.
S. lycopersicoides ILs (97). This group of ILs have been bred from S. lycopersicoides LA2951 into the background of S. lycopersicum cv. VF36. These lines represent ~96% of the donor genome and are described in Canady et al. 2005 Genome 48:685, and Chetelat and Meglic 2000 Theor. Appl. Genet. 76:647. While some lines are available in the homozygous condition, others are partially or completely sterile as homozygotes, thus are maintained via heterozygotes. In this case, marker analysis is required to identify the desired genotypes in segregating progenies. Seed of some lines may be limited or temporarily unavailable. Click here for more information.
Backcross Recombinant Inbreds (90). This group of backcross recombinant inbred lines originated from the cross S. lycopersicum cv. E-6203 × S. pimpinellifolium LA1589 (Genome 45:1189). The result of 2 BC’s and at least 6 generations of single seed descent, the lines are highly homozygous (residual heterozygosity ~3%). The population has been genotyped at 127 marker loci, and the corresponding maps, map files, and QTL data are available from the Solanaceae Genomics Network. This set of 90 lines has been selected for optimum mapping resolution using the MapPop software, and provide a permanent, high resolution mapping population.
Recombinant Inbred Lines (148). A set of RILs from S. lycopersicum cv. NC-EBR1 x S. pimpinellifolium LA2093 was developed by Ashrafi et al. 2009 Genome 52: 935. The population has been genotyped to very high resolution via GBS, as reported by Gonda et al. 2019 The Plant Genome 12: 180010.
S. pennellii Backcross Inbred Lines (435 + 53). This collection of BILs was developed by Ofner et al 2016 (Plant Journal 87: 151) from the cross S. lycopersicum cv. M-82 x S. pennellii LA0716. The population has been genotyped to high resolution using GBS by Fulop et al. 2016 (G3 Genes/Genomes/Genetics 6: 3169). Another set of BILs was developed using the S. pennellii accession LA5240, aka the "LOST" accession, backcrossed into the background of cv. LEA, as described by Torgeman & Zamir 2023 PNAS and Torgeman et al. 2024 Plant J. The TGRC maintains a subset of 53 LOST BILs, selected to provide maximum genome coverage. Marker data on the LOST BILs subset can be downloaded here. To view a list of BILs, click on Database Queries/Core Collections and Mapping Populations, then choose the corresponding accession group.
Alien Substitution Lines (10). In the course of his study of segregation and recombination in S. lycopersicum x S. pennellii hybrids, Rick (1969 Genetics 26:753; and 1972 Biol. Zbl. 91:209) backcrossed several chromosomes from S. pennellii LA0716 into the background of the corresponding chromosome marker stocks in S. lycopersicum. Selected heterozygotes of later generations were selfed and progenies containing the wild type alleles at the marker loci were selected. The chromosome 6 substitution (LA3142) was further selected with RFLP markers to eliminate residual heterozygosity (Weide et al. 1993 Genetics 135:1175). The mutant loci used to select each substitution are indicated. In addition, three chromosome substitution lines (SL-7, -8 and -10) representing S. lycopersicoides LA2951 chromosomes S. lycopersicum cv. VF36 are included here. These are all maintained as heterozygotes, as described by Ji and Chetelat 2003 (Theor. Appl. Genet. 106: 979).
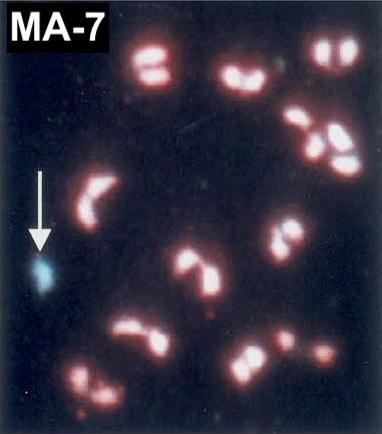
Monosomic Alien Addition Lines (10). A partial set of monosomic addition lines (MAs), each containing a single extra chromosome from S. lycopersicoides LA1964 added to the genome of cultivated tomato was developed by Chetelat et al. 1998 (Genome 41:40). The extra chromosomes pair and recombine with their tomato homeologues at varying rates, as described by Ji and Chetelat 2003 (Theor Appl Genet 106:979). The integrity of the S. lycopersicoides chromosomes in these stocks has been tested with a limited number of markers, hence some may be recombinant. For example, our stock of MA-8 lacks S. lycopersicoides markers distal to TG330 on the long arm. Furthermore, we were unable to maintain MA-1 and MA-6, both of which are now extinct. Like other types of trisomics, progeny of the monosomic additions include both diploids and trisomics, the proportion of which varies between each chromosome group. Identification of monosomic additions in each generation is facilitated by their phenotypic resemblance to the corresponding primary trisomic. Therefore, our guidelines for identifying trisomics in the seedling stage are useful for selecting monosomic additions as well.
Other Prebreds (30). This group of prebreds contain selected morphological traits bred into cultivated tomato from related wild species. Some traits may be simply inherited, others likely involve multiple genetic loci.
Stress Tolerant Stocks (44+). We receive many requests for stocks with tolerances to abiotic stresses. Therefore, we chose this group of mostly wild species accessions based on our observations of plants in their native habitats and/or reports in the literature.
Translocations (38). Our translocation stocks have been assembled from the collections of their originators - D.W. Barton, C.D. Clayberg, B.S. Gill, G.R. Stringham, B. Snoad, and G. Khush. As far as we know, they are all homozygous for the indicated structural changes. They are described by Gill et al. (TGC Report 23: 17-18; TGC Report 24:10-12). Accessions with an asterisk comprise the tester set.
Trisomics (35). We maintain a series of trisomics containing various kinds of extra chromosomes. Since the extras are transmitted irregularly, each stock necessarily produce a majority of diploid progeny, the remainder aneuploid. Primary trisomics yield mostly 2n and 2n+1, and rarely tetrasomics (2n+2). Telotrisomics yield telos and an occasional rare tetratelosomic. Secondary, tertiary, and compensating trisomics transmit other trisomic types as expected. Because transmission is irregular and reproduction of stocks requires much labor, our stocks are limited. In requesting our aneuploids, researchers are asked to keep these points in mind. To assist in the identification of primary trisomics at the seedling stage, the distinguishing morphological features of each trisomic have been summarized by Rick (TGC Report 37:60). See papers by Khush and Rick for descriptions of the telotrisomics, secondary trisomics, tertiary trisomics, and compensating trisomics. Additional 2n+1 stocks are listed under Monosomic Alien Addition Lines.
Autotetraploids (16). We maintain tetraploid stocks of S. lycopersicum and several wild relatives. Some of the tetraploid S. lycopersicum originated from spontaneous chromosome doubling, others were induced.

Cytoplasmic Variants (3). These lines are cytoplasmically-inherited chlorotic variants and are included in the miscellaneous group for want of better classification. They were induced by mutagens and are inherited in a strictly maternal fashion. They are not transmitted through pollen but in reciprocal crosses -- no matter what male parents we have used -- the progeny are 100% variant.
Chromosome Marker Stocks (178). This group consists of stocks containing a series of marker genes representing a single chromosome. In a few cases markers on other chromosomes are also present (listed in parentheses). Some of the more useful stocks have been combined with male steriles in order to facilitate large scale test crossing. These marker stocks are grouped by chromosome, with asterisks indicating the preferred genotype, i.e. that which provides provides the most complete map coverage for linkage tests.
Linkage Screening Testers (15). These linkage tester stocks each combine two pairs of strategically situated markers on two different chromosomes (see Rick and Zobel 1972 TGC Report 22: 24). They are intended primarily for assigning new, unmapped markers to a chromosome. The more complete chromosome marker combinations (list 6.1 above) would typically be used for subsequent testing to delimit loci more accurately. Whereas six of these stocks should pretty well cover the tomato genome, the list includes the entire series of the current available testers because alternative stocks differ in their usefulness, depending upon the phenotype of the new mutant to be located. The chromosomal location of each pair of markers is indicated in parentheses.
Miscellaneous Marker Combinations (305). This group includes multiple marker stocks in which various mutant genes, not necessarily on the same chromosome, have been combined. A few of these lines contain linked genes but are classified here because other linkage tester stocks provide the same combinations or because they are more useful as markers of several chromosomes. Some multiple marker combinations that are of limited usefulness, difficult to maintain, and/or redundant with other genotypes, have been dropped from the current list.
